遗传算法遗传算法(GeneticAlgorithm)是模拟达尔文生物进化论的自然选择和遗传学机理的生物进化过程的计算模型,是一种通过模拟自然进化过程搜索最优解的方法,它最初由美国Michigan大学J.Holland教授于1975年首先提出来的,并出版了颇有影响的专著《Adaptationin Natural and ArtificialSystems》,GA这个名称才逐渐为人所知,J.Holland教授所提出的GA通常为简单遗传算法(SGA)。遗传算法的MATLAB程序%遗传算法程序:说明:fga.m 为遗传算法的主程序; 采用二进制Gray编码,采用基于轮盘赌法的非线性排名选择,均匀交叉,变异操作,而且还引入了倒位操作!function[BestPop,Trace]=fga(FUN,LB,UB,eranum,popsize,pCross,pMutation,pInversion,options)
%[BestPop,Trace]=fmaxga(FUN,LB,UB,eranum,popsize,pcross,pmutation)
% Finds a??maximum of afunction of several variables.
% fmaxga solves problemsof the form:
% max F(X) subject to:LB <= X <=UB
%BestPop - 最优的群体即为最优的染色体群
% Trace -最佳染色体所对应的目标函数值
% FUN - 目标函数
% LB -自变量下限
% UB -自变量上限
%eranum -种群的代数,取100--1000(默认200)
%popsize -每一代种群的规模;此可取50--200(默认100)
%pcross - 交叉概率,一般取0.5--0.85之间较好(默认0.8)
%pmutation -初始变异概率,一般取0.05-0.2之间较好(默认0.1)
%pInversion -倒位概率,一般取0.05-0.3之间较好(默认0.2)
%options -1*2矩阵,options(1)=0二进制编码(默认0),option(1)~=0十进制编码,option(2)设定求解精度(默认1e-4)
T1=clock;
ifnargin<3, error('FMAXGA requires at least threeinput arguments'); end
if nargin==3,eranum=200;popsize=100;pCross=0.8;pMutation=0.1;pInversion=0.15;options=[01e-4];end
if nargin==4,popsize=100;pCross=0.8;pMutation=0.1;pInversion=0.15;options=[01e-4];end
if nargin==5,pCross=0.8;pMutation=0.1;pInversion=0.15;options=[01e-4];end
if nargin==6,pMutation=0.1;pInversion=0.15;options=[01e-4];end
if nargin==7,pInversion=0.15;options=[0 1e-4];end
iffind((LB-UB)>0)
error('数据输入错误,请重新输入(LB<UB):');
end
s=sprintf('程序运行需要约%.4f秒钟时间,请稍等......',(eranum*popsize/1000));
disp(s);
global m n NewPopchildren1 children2 VarNum
bounds=[LB;UB]';bits=[];VarNum=size(bounds,1);
precision=options(2);%由求解精度确定二进制编码长度
bits=ceil(log2((bounds(:,2)-bounds(:,1))'./ precision));%由设定精度划分区间
[Pop]=InitPopGray(popsize,bits);%初始化种群
[m,n]=size(Pop);
NewPop=zeros(m,n);
children1=zeros(1,n);
children2=zeros(1,n);
pm0=pMutation;
BestPop=zeros(eranum,n);%分配初始解空间BestPop,Trace
Trace=zeros(eranum,length(bits)+1);
i=1;
whilei<=eranum
for j=1:m
value(j)=feval_r(FUN(1,:),(b2f(Pop(j,:),bounds,bits)));%计算适应度
end
[MaxValue,Index]=max(value);
BestPop(i,:)=Pop(Index,:);
Trace(i,1)=MaxValue;
Trace(i,(2:length(bits)+1))=b2f(BestPop(i,:),bounds,bits);
[selectpop]=NonlinearRankSelect(FUN,Pop,bounds,bits);%非线性排名选择
[CrossOverPop]=CrossOver(selectpop,pCross,round(unidrnd(eranum-i)/eranum));%采用多点交叉和均匀交叉,且逐步增大均匀交叉的概率
round(unidrnd(eranum-i)/eranum)
[MutationPop]=Mutation(CrossOverPop,pMutation,VarNum);%变异
[InversionPop]=Inversion(MutationPop,pInversion);%倒位
Pop=InversionPop;%更新
pMutation=pm0+(i^4)*(pCross/3-pm0)/(eranum^4);
%随着种群向前进化,逐步增大变异率至1/2交叉率
p(i)=pMutation;
i=i+1;
end
t=1:eranum;
plot(t,Trace(:,1)');
title('函数优化的遗传算法');xlabel('进化世代数(eranum)');ylabel('每一代最优适应度(maxfitness)');
[MaxFval,I]=max(Trace(:,1));
X=Trace(I,(2:length(bits)+1));
holdon;??plot(I,MaxFval,'*');
text(I+5,MaxFval,['FMAX='num2str(MaxFval)]);
str1=sprintf('进化到 %d 代,自变量为 %s 时,得本次求解的最优值%fn对应染色体是:%s',I,num2str(X),MaxFval,num2str(BestPop(I,:)));
disp(str1);
%figure(2);plot(t,p);%绘制变异值增大过程
T2=clock;
elapsed_time=T2-T1;
ifelapsed_time(6)<0
elapsed_time(6)=elapsed_time(6)+60;elapsed_time(5)=elapsed_time(5)-1;
end
ifelapsed_time(5)<0
elapsed_time(5)=elapsed_time(5)+60;elapsed_time(4)=elapsed_time(4)-1;
end %像这种程序当然不考虑运行上小时啦
str2=sprintf('程序运行耗时 %d小时 %d 分钟 %.4f秒',elapsed_time(4),elapsed_time(5),elapsed_time(6));
disp(str2);
%初始化种群
%采用二进制Gray编码,其目的是为了克服二进制编码的Hamming悬崖缺点
function[initpop]=InitPopGray(popsize,bits)
len=sum(bits);
initpop=zeros(popsize,len);%Thewhole zero encoding individual
fori=2:popsize-1
pop=round(rand(1,len));
pop=mod(([0 pop]+[pop 0]),2);
%i=1时,b(1)=a(1);i>1时,b(i)=mod(a(i-1)+a(i),2)
%其中原二进制串:a(1)a(2)...a(n),Gray串:b(1)b(2)...b(n)
initpop(i,:)=pop(1:end-1);
end
initpop(popsize,:)=ones(1,len);%Thewhole one encoding individual
%解码
function [fval] =b2f(bval,bounds,bits)
% fval - 表征各变量的十进制数
% bval - 表征各变量的二进制编码串
% bounds -各变量的取值范围
% bits - 各变量的二进制编码长度
scale=(bounds(:,2)-bounds(:,1))'./(2.^bits-1);%The range of the variables
numV=size(bounds,1);
cs=[0cumsum(bits)];
fori=1:numV
a=bval((cs(i)+1):cs(i+1));
fval(i)=sum(2.^(size(a,2)-1:-1:0).*a)*scale(i)+bounds(i,1);
end
%选择操作 %采用基于轮盘赌法的非线性排名选择
%各个体成员按适应值从大到小分配选择概率:
%P(i)=(q/1-(1-q)^n)*(1-q)^i,??其中P(0)>P(1)>...>P(n),sum(P(i))=1
function[selectpop]=NonlinearRankSelect(FUN,pop,bounds,bits)
global mn
selectpop=zeros(m,n);
fit=zeros(m,1);
fori=1:m
fit(i)=feval_r(FUN(1,:),(b2f(pop(i,:),bounds,bits)));%以函数值为适应值做排名依据
end
selectprob=fit/sum(fit);%计算各个体相对适应度(0,1)
q=max(selectprob);%选择最优的概率
x=zeros(m,2);
x(:,1)=[m:-1:1]';
[yx(:,2)]=sort(selectprob);
r=q/(1-(1-q)^m);%标准分布基值
newfit(x(:,2))=r*(1-q).^(x(:,1)-1);%生成选择概率
newfit=cumsum(newfit);%计算各选择概率之和
rNums=sort(rand(m,1));
fitIn=1;newIn=1;
whilenewIn<=m
ifrNums(newIn)<newfit(fitIn)
selectpop(newIn,:)=pop(fitIn,:);
newIn=newIn+1;
else
fitIn=fitIn+1;
end
end
%交叉操作
function[NewPop]=CrossOver(OldPop,pCross,opts)
%OldPop为父代种群,pcross为交叉概率
global m nNewPop
r=rand(1,m);
y1=find(r<pCross);
y2=find(r>=pCross);
len=length(y1);
iflen>2&mod(len,2)==1%如果用来进行交叉的染色体的条数为奇数,将其调整为偶数
y2(length(y2)+1)=y1(len);
y1(len)=[];
end
iflength(y1)>=2
for i=0:2:length(y1)-2
ifopts==0
[NewPop(y1(i+1),:),NewPop(y1(i+2),:)]=EqualCrossOver(OldPop(y1(i+1),:),OldPop(y1(i+2),:));
else
[NewPop(y1(i+1),:),NewPop(y1(i+2),:)]=MultiPointCross(OldPop(y1(i+1),:),OldPop(y1(i+2),:));
end
end
end
NewPop(y2,:)=OldPop(y2,:);
%采用均匀交叉
function[children1,children2]=EqualCrossOver(parent1,parent2)
global n children1children2
hidecode=round(rand(1,n));%随机生成掩码
crossposition=find(hidecode==1);
holdposition=find(hidecode==0);
children1(crossposition)=parent1(crossposition);%掩码为1,父1为子1提供基因
children1(holdposition)=parent2(holdposition);%掩码为0,父2为子1提供基因
children2(crossposition)=parent2(crossposition);%掩码为1,父2为子2提供基因
children2(holdposition)=parent1(holdposition);%掩码为0,父1为子2提供基因
%采用多点交叉,交叉点数由变量数决定
function[Children1,Children2]=MultiPointCross(Parent1,Parent2)
global n Children1Children2 VarNum
Children1=Parent1;
Children2=Parent2;
Points=sort(unidrnd(n,1,2*VarNum));
fori=1:VarNum
Children1(Points(2*i-1):Points(2*i))=Parent2(Points(2*i-1):Points(2*i));
Children2(Points(2*i-1):Points(2*i))=Parent1(Points(2*i-1):Points(2*i));
end
%变异操作
function[NewPop]=Mutation(OldPop,pMutation,VarNum)
global m nNewPop
r=rand(1,m);
position=find(r<=pMutation);
len=length(position);
iflen>=1
for i=1:len
k=unidrnd(n,1,VarNum);%设置变异点数,一般设置1点
forj=1:length(k)
ifOldPop(position(i),k(j))==1
OldPop(position(i),k(j))=0;
else
OldPop(position(i),k(j))=1;
end
end
end
end
NewPop=OldPop;
%倒位操作
function[NewPop]=Inversion(OldPop,pInversion)
global m nNewPop
NewPop=OldPop;
r=rand(1,m);
PopIn=find(r<=pInversion);
len=length(PopIn);
iflen>=1
for i=1:len
d=sort(unidrnd(n,1,2));
ifd(1)~=1&d(2)~=n
NewPop(PopIn(i),1:d(1)-1)=OldPop(PopIn(i),1:d(1)-1);
NewPop(PopIn(i),d(1):d(2))=OldPop(PopIn(i),d(2 ):-1:d(1));
NewPop(PopIn(i),d(2)+1:n)=OldPop(PopIn(i),d(2)+1:n);
end
end
end
%遗传算法程序:说明: fga.m为遗传算法的主程序; 采用二进制Gray编码,采用基于轮盘赌法的非线性排名选择,均匀交叉,变异操作,而且还引入了倒位操作!
function[BestPop,Trace]=fga(FUN,LB,UB,eranum,popsize,pCross,pMutation,pInversion,options)
%[BestPop,Trace]=fmaxga(FUN,LB,UB,eranum,popsize,pcross,pmutation)
% Finds a??maximum of afunction of several variables.
% fmaxga solves problemsof the form:
% max F(X) subject to:LB <= X <=UB
%BestPop - 最优的群体即为最优的染色体群
% Trace -最佳染色体所对应的目标函数值
% FUN - 目标函数
% LB -自变量下限
% UB -自变量上限
%eranum -种群的代数,取100--1000(默认200)
%popsize -每一代种群的规模;此可取50--200(默认100)
%pcross - 交叉概率,一般取0.5--0.85之间较好(默认0.8)
%pmutation -初始变异概率,一般取0.05-0.2之间较好(默认0.1)
%pInversion -倒位概率,一般取0.05-0.3之间较好(默认0.2)
%options -1*2矩阵,options(1)=0二进制编码(默认0),option(1)~=0十进制编码,option(2)设定求解精度(默认1e-4)
T1=clock;
ifnargin<3, error('FMAXGA requires at least threeinput arguments'); end
if nargin==3,eranum=200;popsize=100;pCross=0.8;pMutation=0.1;pInversion=0.15;options=[01e-4];end
if nargin==4,popsize=100;pCross=0.8;pMutation=0.1;pInversion=0.15;options=[01e-4];end
if nargin==5,pCross=0.8;pMutation=0.1;pInversion=0.15;options=[01e-4];end
if nargin==6,pMutation=0.1;pInversion=0.15;options=[01e-4];end
if nargin==7,pInversion=0.15;options=[0 1e-4];end
iffind((LB-UB)>0)
error('数据输入错误,请重新输入(LB<UB):');
end
s=sprintf('程序运行需要约%.4f秒钟时间,请稍等......',(eranum*popsize/1000));
disp(s);
global m n NewPopchildren1 children2 VarNum
bounds=[LB;UB]';bits=[];VarNum=size(bounds,1);
precision=options(2);%由求解精度确定二进制编码长度
bits=ceil(log2((bounds(:,2)-bounds(:,1))'./ precision));%由设定精度划分区间
[Pop]=InitPopGray(popsize,bits);%初始化种群
[m,n]=size(Pop);
NewPop=zeros(m,n);
children1=zeros(1,n);
children2=zeros(1,n);
pm0=pMutation;
BestPop=zeros(eranum,n);%分配初始解空间BestPop,Trace
Trace=zeros(eranum,length(bits)+1);
i=1;
whilei<=eranum
for j=1:m
value(j)=feval_r(FUN(1,:),(b2f(Pop(j,:),bounds,bits)));%计算适应度
end
[MaxValue,Index]=max(value);
BestPop(i,:)=Pop(Index,:);
Trace(i,1)=MaxValue;
Trace(i,(2:length(bits)+1))=b2f(BestPop(i,:),bounds,bits);
[selectpop]=NonlinearRankSelect(FUN,Pop,bounds,bits);%非线性排名选择
[CrossOverPop]=CrossOver(selectpop,pCross,round(unidrnd(eranum-i)/eranum));%采用多点交叉和均匀交叉,且逐步增大均匀交叉的概率
round(unidrnd(eranum-i)/eranum)
[MutationPop]=Mutation(CrossOverPop,pMutation,VarNum);%变异
[InversionPop]=Inversion(MutationPop,pInversion);%倒位
Pop=InversionPop;%更新
pMutation=pm0+(i^4)*(pCross/3-pm0)/(eranum^4);
%随着种群向前进化,逐步增大变异率至1/2交叉率
p(i)=pMutation;
i=i+1;
end
t=1:eranum;
plot(t,Trace(:,1)');
title('函数优化的遗传算法');xlabel('进化世代数(eranum)');ylabel('每一代最优适应度(maxfitness)');
[MaxFval,I]=max(Trace(:,1));
X=Trace(I,(2:length(bits)+1));
holdon;??plot(I,MaxFval,'*');
text(I+5,MaxFval,['FMAX='num2str(MaxFval)]);
str1=sprintf('进化到 %d 代,自变量为 %s 时,得本次求解的最优值%fn对应染色体是:%s',I,num2str(X),MaxFval,num2str(BestPop(I,:)));
disp(str1);
%figure(2);plot(t,p);%绘制变异值增大过程
T2=clock;
elapsed_time=T2-T1;
ifelapsed_time(6)<0
elapsed_time(6)=elapsed_time(6)+60;elapsed_time(5)=elapsed_time(5)-1;
end
ifelapsed_time(5)<0
elapsed_time(5)=elapsed_time(5)+60;elapsed_time(4)=elapsed_time(4)-1;
end %像这种程序当然不考虑运行上小时啦
str2=sprintf('程序运行耗时 %d小时 %d 分钟 %.4f秒',elapsed_time(4),elapsed_time(5),elapsed_time(6));
disp(str2);
%初始化种群
%采用二进制Gray编码,其目的是为了克服二进制编码的Hamming悬崖缺点
function[initpop]=InitPopGray(popsize,bits)
len=sum(bits);
initpop=zeros(popsize,len);%Thewhole zero encoding individual
fori=2:popsize-1
pop=round(rand(1,len));
pop=mod(([0 pop]+[pop 0]),2);
%i=1时,b(1)=a(1);i>1时,b(i)=mod(a(i-1)+a(i),2)
%其中原二进制串:a(1)a(2)...a(n),Gray串:b(1)b(2)...b(n)
initpop(i,:)=pop(1:end-1);
end
initpop(popsize,:)=ones(1,len);%Thewhole one encoding individual
%解码
function [fval] =b2f(bval,bounds,bits)
% fval - 表征各变量的十进制数
% bval - 表征各变量的二进制编码串
% bounds -各变量的取值范围
% bits - 各变量的二进制编码长度
scale=(bounds(:,2)-bounds(:,1))'./(2.^bits-1);%The range of the variables
numV=size(bounds,1);
cs=[0cumsum(bits)];
fori=1:numV
a=bval((cs(i)+1):cs(i+1));
fval(i)=sum(2.^(size(a,2)-1:-1:0).*a)*scale(i)+bounds(i,1);
end
%选择操作 %采用基于轮盘赌法的非线性排名选择
%各个体成员按适应值从大到小分配选择概率:
%P(i)=(q/1-(1-q)^n)*(1-q)^i,??其中P(0)>P(1)>...>P(n),sum(P(i))=1
function[selectpop]=NonlinearRankSelect(FUN,pop,bounds,bits)
global mn
selectpop=zeros(m,n);
fit=zeros(m,1);
fori=1:m
fit(i)=feval_r(FUN(1,:),(b2f(pop(i,:),bounds,bits)));%以函数值为适应值做排名依据
end
selectprob=fit/sum(fit);%计算各个体相对适应度(0,1)
q=max(selectprob);%选择最优的概率
x=zeros(m,2);
x(:,1)=[m:-1:1]';
[yx(:,2)]=sort(selectprob);
r=q/(1-(1-q)^m);%标准分布基值
newfit(x(:,2))=r*(1-q).^(x(:,1)-1);%生成选择概率
newfit=cumsum(newfit);%计算各选择概率之和
rNums=sort(rand(m,1));
fitIn=1;newIn=1;
whilenewIn<=m
ifrNums(newIn)<newfit(fitIn)
selectpop(newIn,:)=pop(fitIn,:);
newIn=newIn+1;
else
fitIn=fitIn+1;
end
end
%交叉操作
function[NewPop]=CrossOver(OldPop,pCross,opts)
%OldPop为父代种群,pcross为交叉概率
global m nNewPop
r=rand(1,m);
y1=find(r<pCross);
y2=find(r>=pCross);
len=length(y1);
iflen>2&mod(len,2)==1%如果用来进行交叉的染色体的条数为奇数,将其调整为偶数
y2(length(y2)+1)=y1(len);
y1(len)=[];
end
iflength(y1)>=2
for i=0:2:length(y1)-2
ifopts==0
[NewPop(y1(i+1),:),NewPop(y1(i+2),:)]=EqualCrossOver(OldPop(y1(i+1),:),OldPop(y1(i+2),:));
else
[NewPop(y1(i+1),:),NewPop(y1(i+2),:)]=MultiPointCross(OldPop(y1(i+1),:),OldPop(y1(i+2),:));
end
end
end
NewPop(y2,:)=OldPop(y2,:);
%采用均匀交叉
function[children1,children2]=EqualCrossOver(parent1,parent2)
global n children1children2
hidecode=round(rand(1,n));%随机生成掩码
crossposition=find(hidecode==1);
holdposition=find(hidecode==0);
children1(crossposition)=parent1(crossposition);%掩码为1,父1为子1提供基因
children1(holdposition)=parent2(holdposition);%掩码为0,父2为子1提供基因
children2(crossposition)=parent2(crossposition);%掩码为1,父2为子2提供基因
children2(holdposition)=parent1(holdposition);%掩码为0,父1为子2提供基因
%采用多点交叉,交叉点数由变量数决定
function[Children1,Children2]=MultiPointCross(Parent1,Parent2)
global n Children1Children2 VarNum
Children1=Parent1;
Children2=Parent2;
Points=sort(unidrnd(n,1,2*VarNum));
fori=1:VarNum
Children1(Points(2*i-1):Points(2*i))=Parent2(Points(2*i-1):Points(2*i));
Children2(Points(2*i-1):Points(2*i))=Parent1(Points(2*i-1):Points(2*i));
end
%变异操作
function[NewPop]=Mutation(OldPop,pMutation,VarNum)
global m nNewPop
r=rand(1,m);
position=find(r<=pMutation);
len=length(position);
iflen>=1
for i=1:len
k=unidrnd(n,1,VarNum);%设置变异点数,一般设置1点
forj=1:length(k)
ifOldPop(position(i),k(j))==1
OldPop(position(i),k(j))=0;
else
OldPop(position(i),k(j))=1;
end
end
end
end
NewPop=OldPop;
%倒位操作
function[NewPop]=Inversion(OldPop,pInversion)
global m nNewPop
NewPop=OldPop;
r=rand(1,m);
PopIn=find(r<=pInversion);
len=length(PopIn);
iflen>=1
for i=1:len
d=sort(unidrnd(n,1,2));
ifd(1)~=1&d(2)~=n
NewPop(PopIn(i),1:d(1)-1)=OldPop(PopIn(i),1:d(1)-1);
NewPop(PopIn(i),d(1):d(2))=OldPop(PopIn(i),d(2):-1:d(1));
NewPop(PopIn(i),d(2)+1:n)=OldPop(PopIn(i),d(2)+1:n);
end
end
end
爱华网本文地址 » http://www.aihuau.com/a/25101016/287302.html
更多阅读
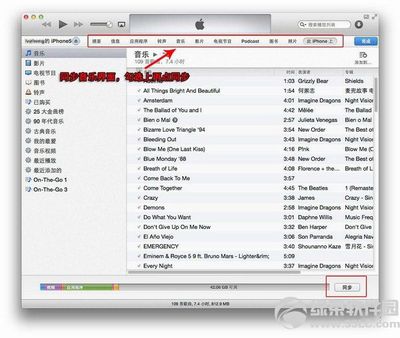
如何用iTunes同步应用程序——简介有了账号,就可以在App Store中下载各种应用和游戏了。那么,如何将iTunes中下载的应用和游戏同步到iPhone中呢?如何用iTunes同步应用程序——方法/步骤如何用iTunes同步应用程序 1、与同步音乐和视频
Floyd最短路算法的MATLAB程序%floyd.m%采用floyd算法计算图a中每对顶点最短路%d是矩离矩阵%r是路由矩阵function[d,r]=floyd(a)n=size(a,1);d=a;fori=1:n

老实说,青岛版第九册《感测技术的应用》这节课让我很头疼啊!像这样讲解型的课,本来就比较难上,学生兴趣不高,所以只能尽量的多安排学生活动来提高学生兴趣。学生情况是农村学生,家里有洗衣机的不多,吸油烟机的不多,也就说身边有传感技术的家

http://blog.chinaunix.net/uid-21016284-id-1831174.htmlBuilding anApplication创建应用程序PS:这部分讲述的是底层的API,当你创建应用程序的时候,你可能使用的是高层的API。#incl
这篇算是一个科普贴,高手绕行。================这里插播一个东西,有些朋友在FQuantStudio公众号上给我留言,我基本都回复了。但有的朋友可能微信设置了关闭接收公众号消息还是什么原因,导致您通过FQuantStudio公众号给我留言,我给您回复

 爱华网
爱华网


